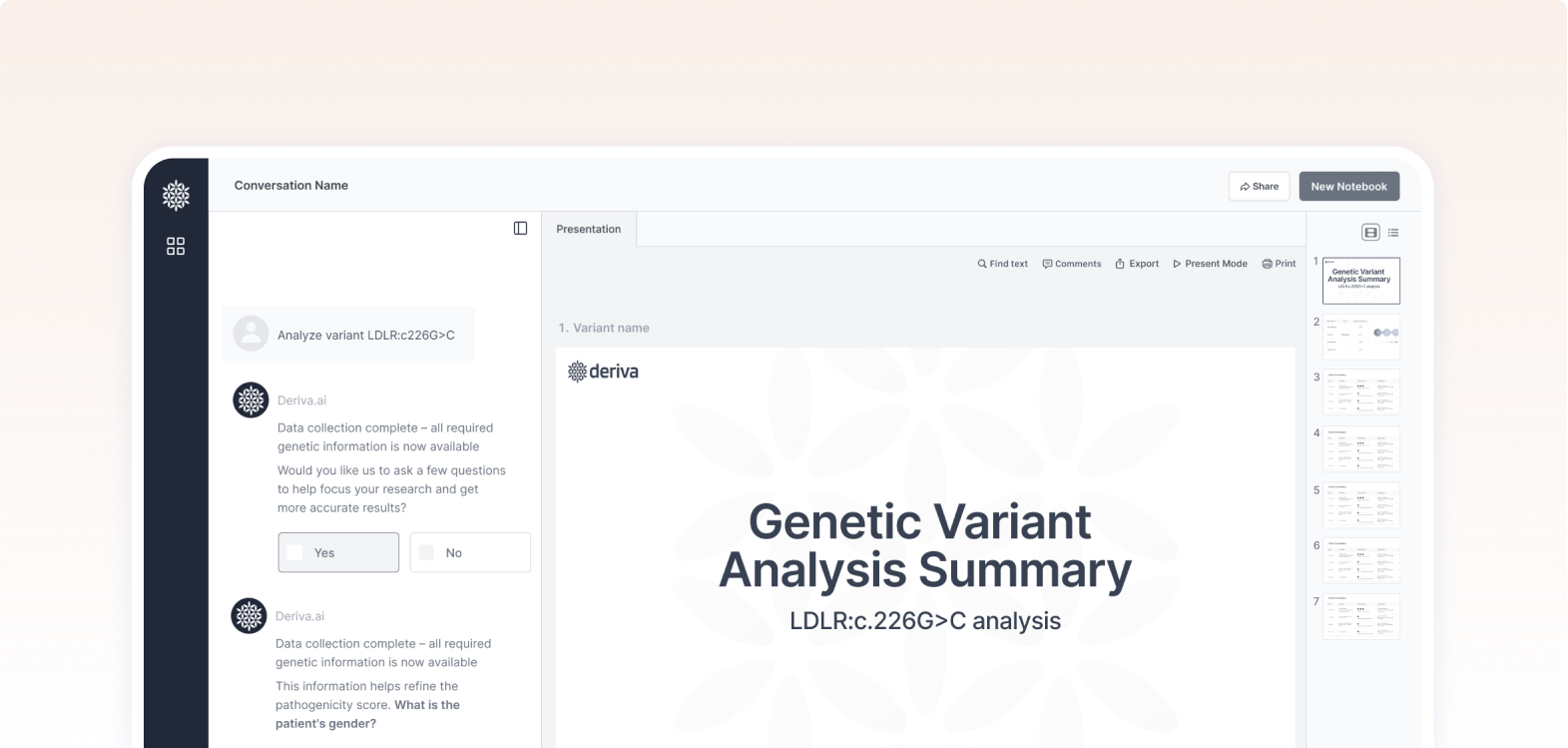
The chat combines open input with structured Yes/No questions to refine results and guide the research process more efficiently.
AI-powered SaaS platform for automating complex research and integrating biological data.
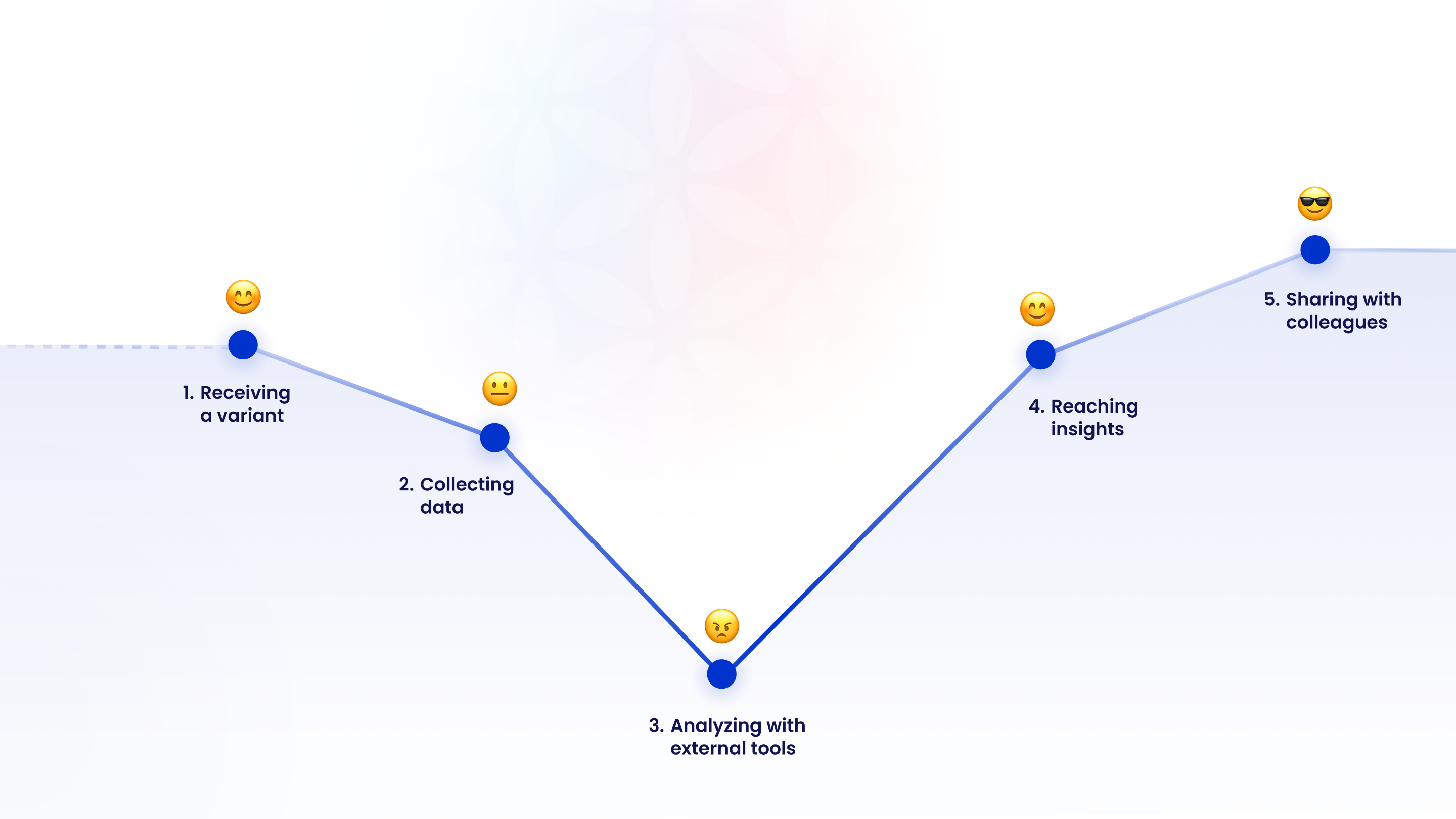
Biology researchers waste valuable time gathering and comparing scattered data instead of focusing on insights. The process is slow, manual, and spread across multiple tools.
Deriva is an AI-based platform that simplifies research by automating data collection, connecting key sources, and helping researchers reach insights faster, with clarity and confidence.
The Challenge of Genetic Research
Every person carries dozens of genetic mutations — most are harmless, but a few may cause disease.
Instead of focusing on insights and discoveries, researchers spend hours searching, comparing, and verifying information.
The core issues
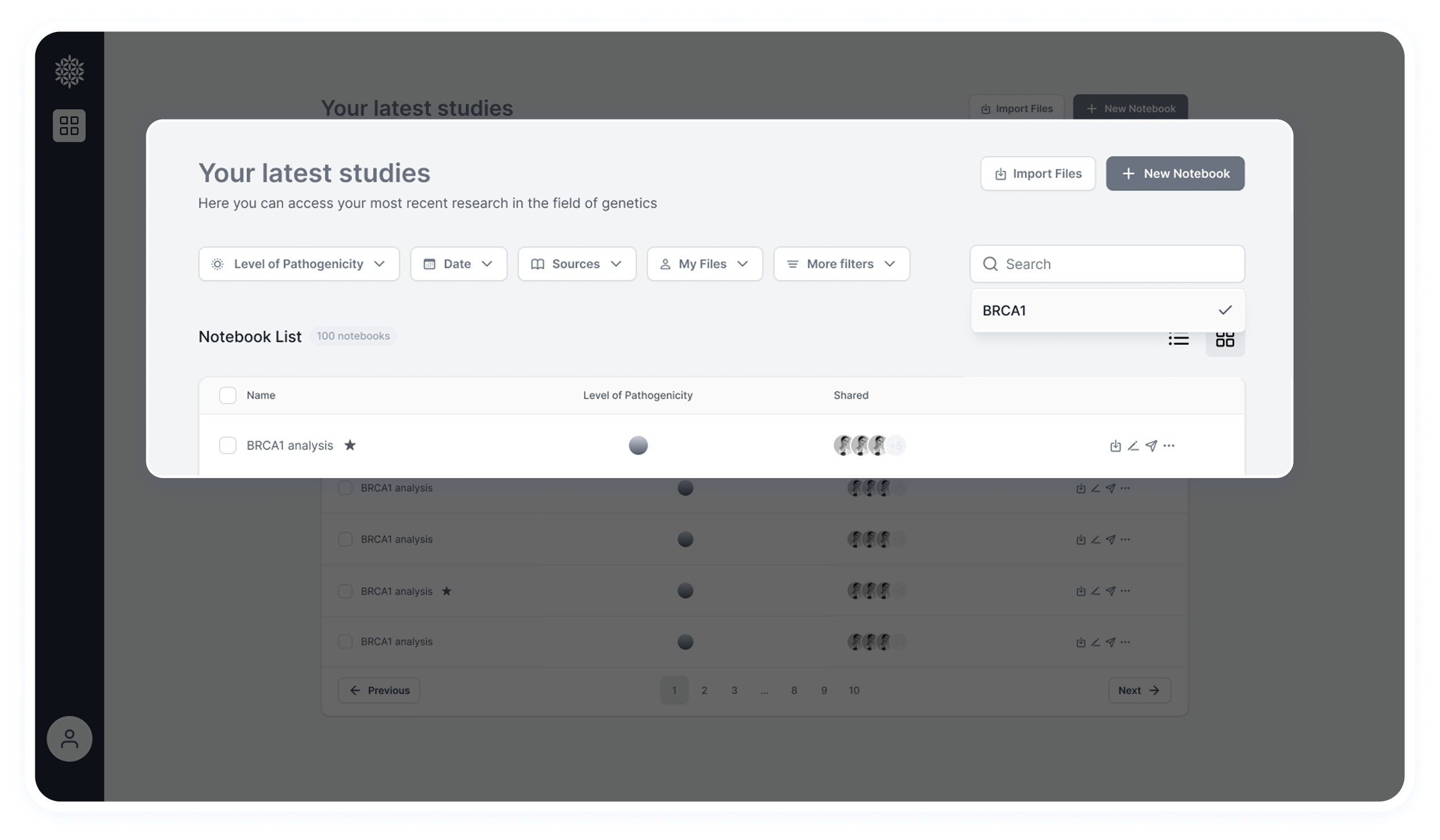
Where Things Stand Today



How Deriva Works
A self-explanatory interface that simplifies complex processes, prioritizes key content, and reduces information overload — giving users clarity and control.
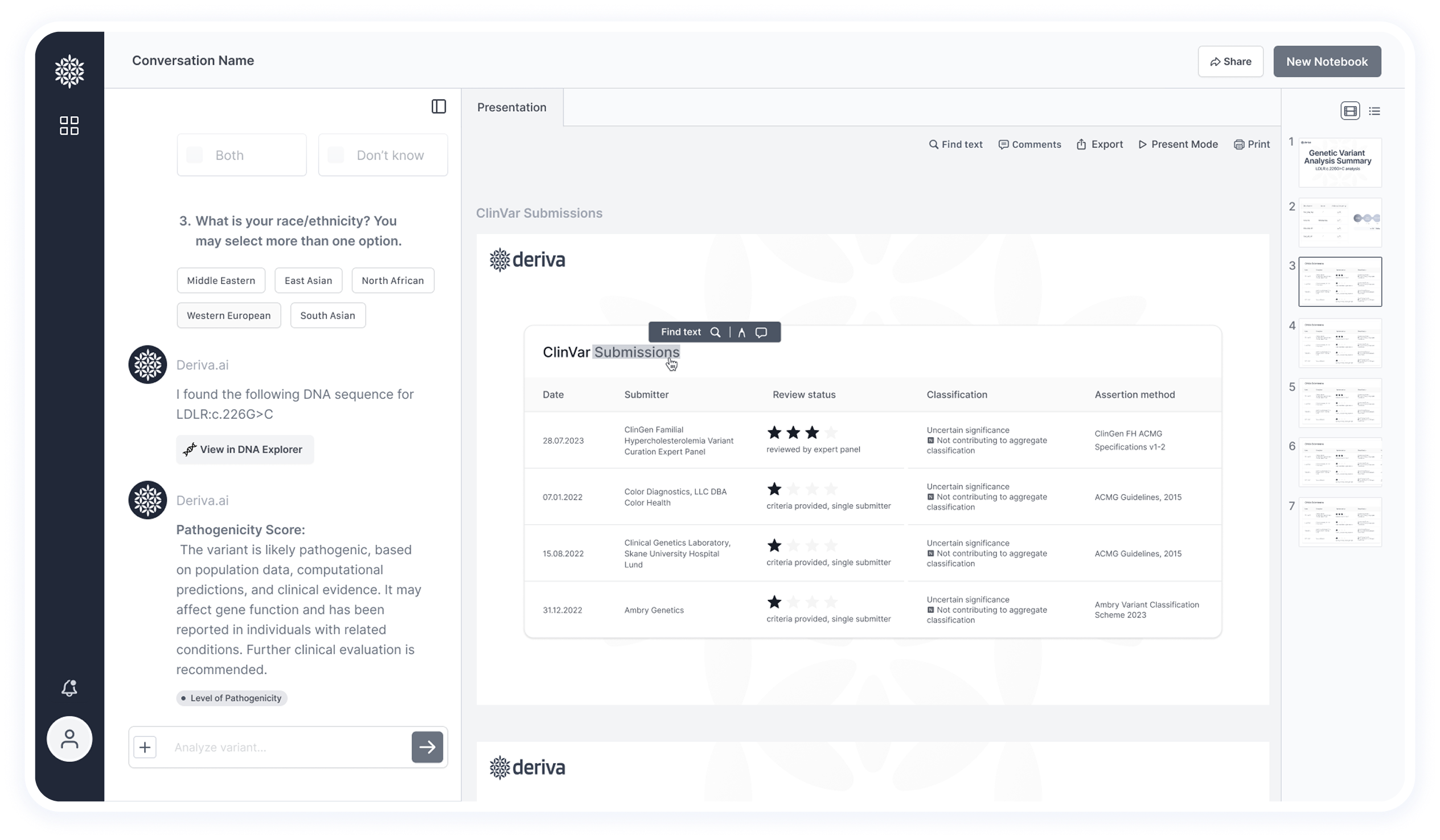
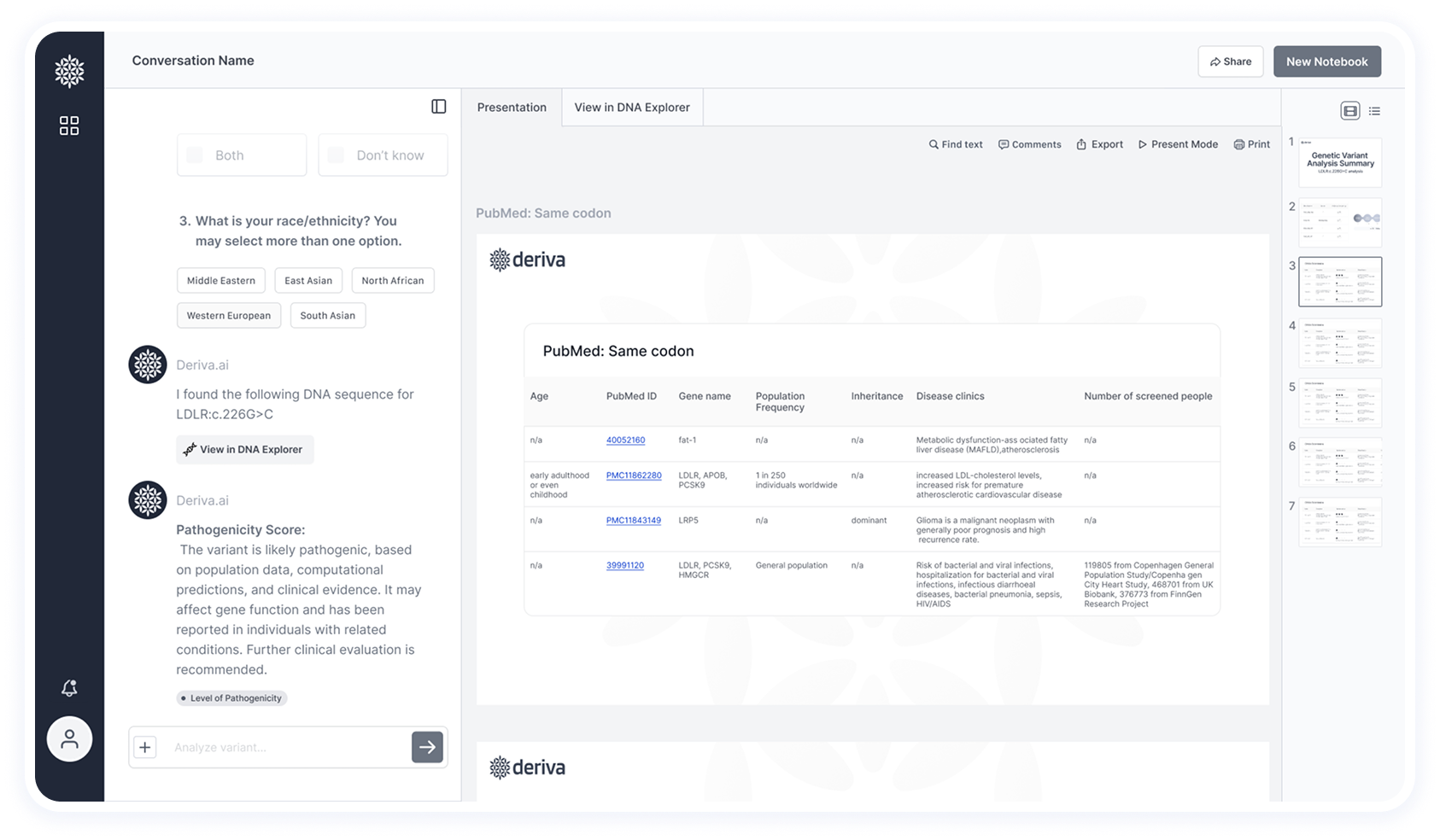
Clear display of trusted sources (PubMed, ClinVar, gnomAD) and traceable reasoning behind results to build confidence.
A natural interface for researchers that mirrors their systematic, source-based workflow — with documentation, cross-referencing, visual tables, and structured exploration.
Seamless research flow with integrated tools — analysis, sources, findings, documentation, and collaboration — all in one platform, no system switching needed.
Encourages full-cycle use with features like history tracking, collaboration, and easy access to past studies.
Highlights relevant data, visualizes findings, and enables quick export — helping researchers reach insights faster.









🗂️ Data Organization and Saving
🧾 Presentation as a Deliverable
🏷️ Tagging and Collaboration
🔔 Alerts and Updates
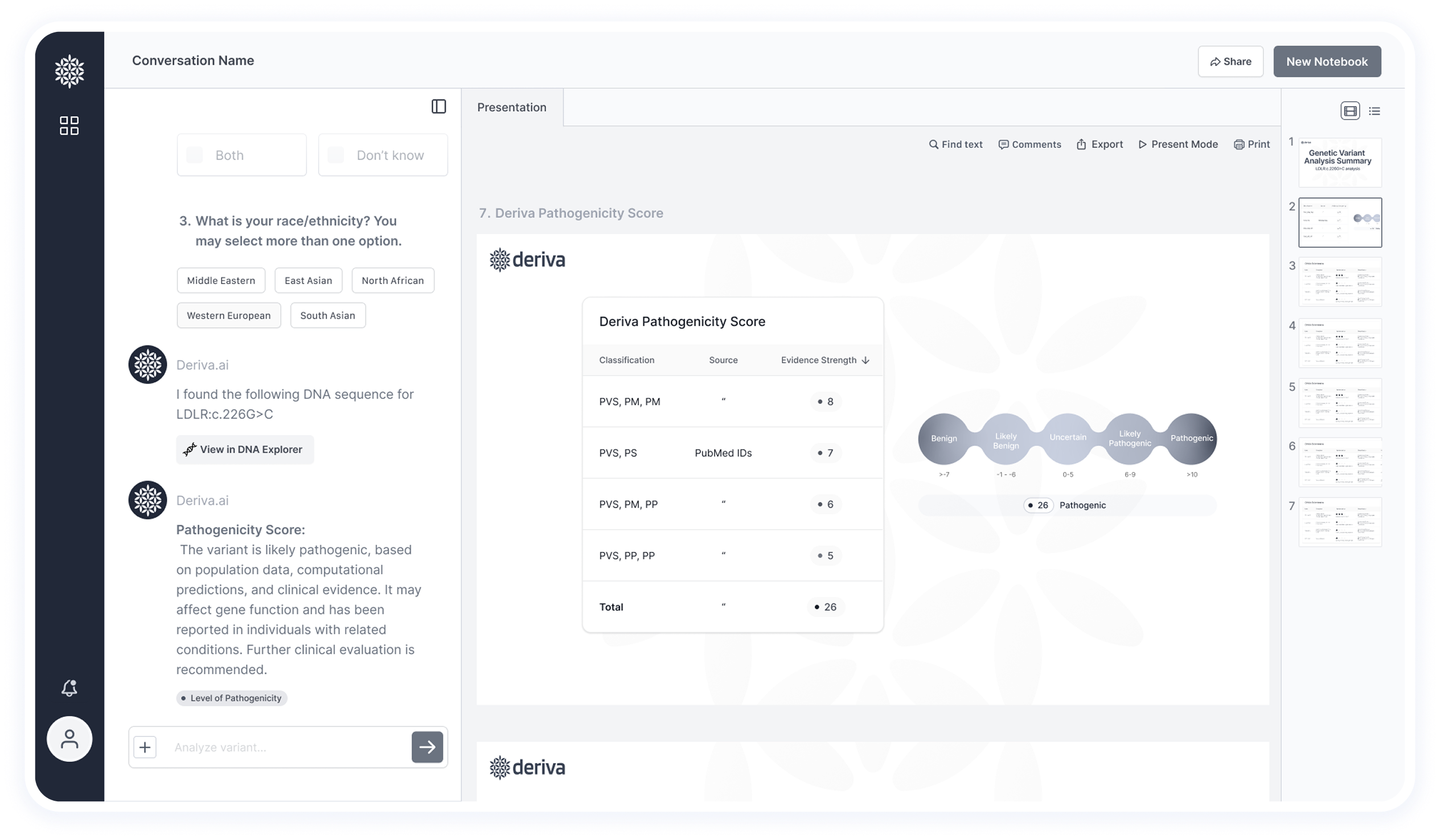
🧬 Pathogenicity-Based Filtering
💻 Desktop-Oriented Design


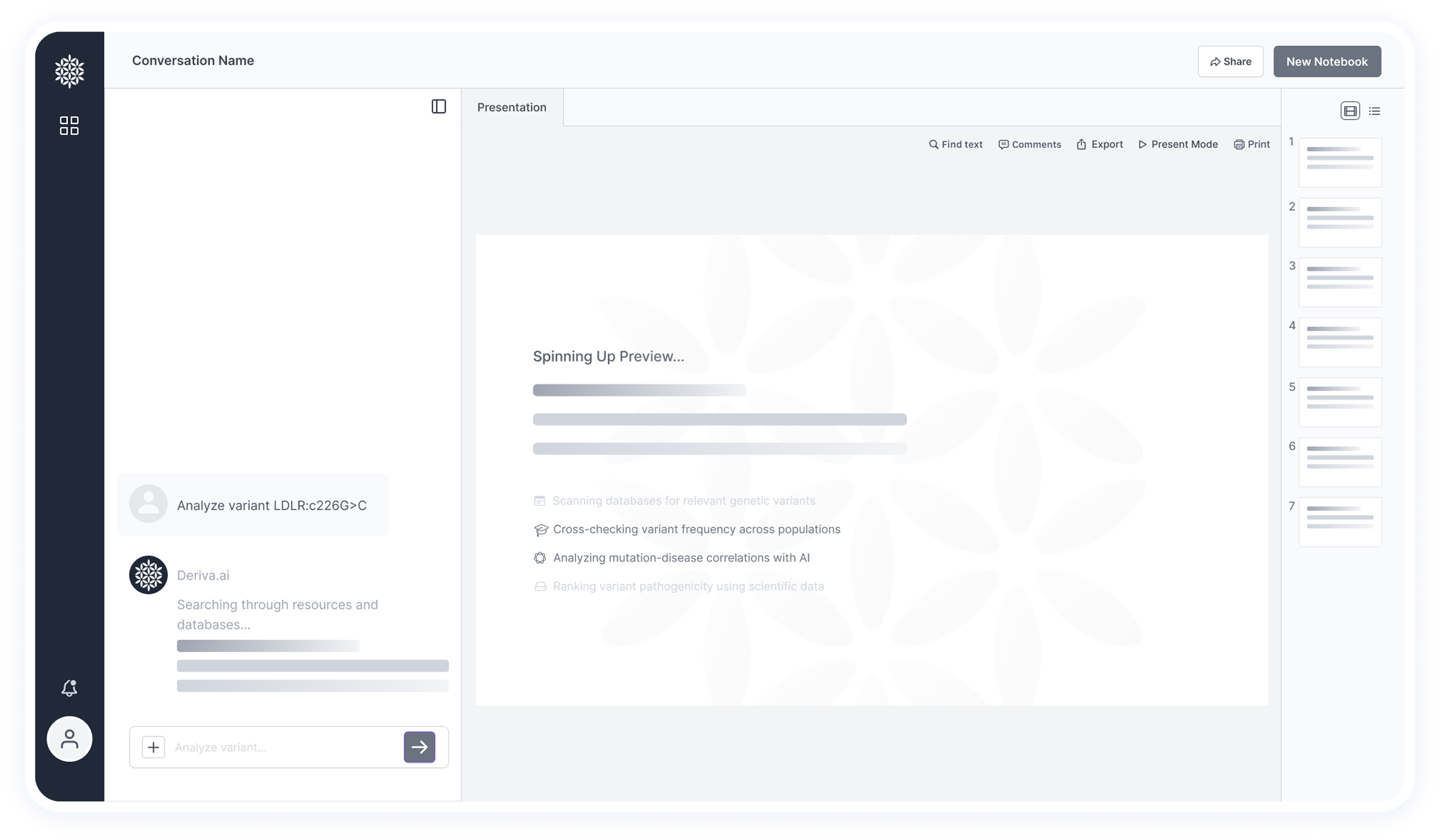
In this part of the interface, we intentionally emphasize the loading and search process.
Showing that the system is actively scanning and retrieving data adds to the sense of transparency and trust
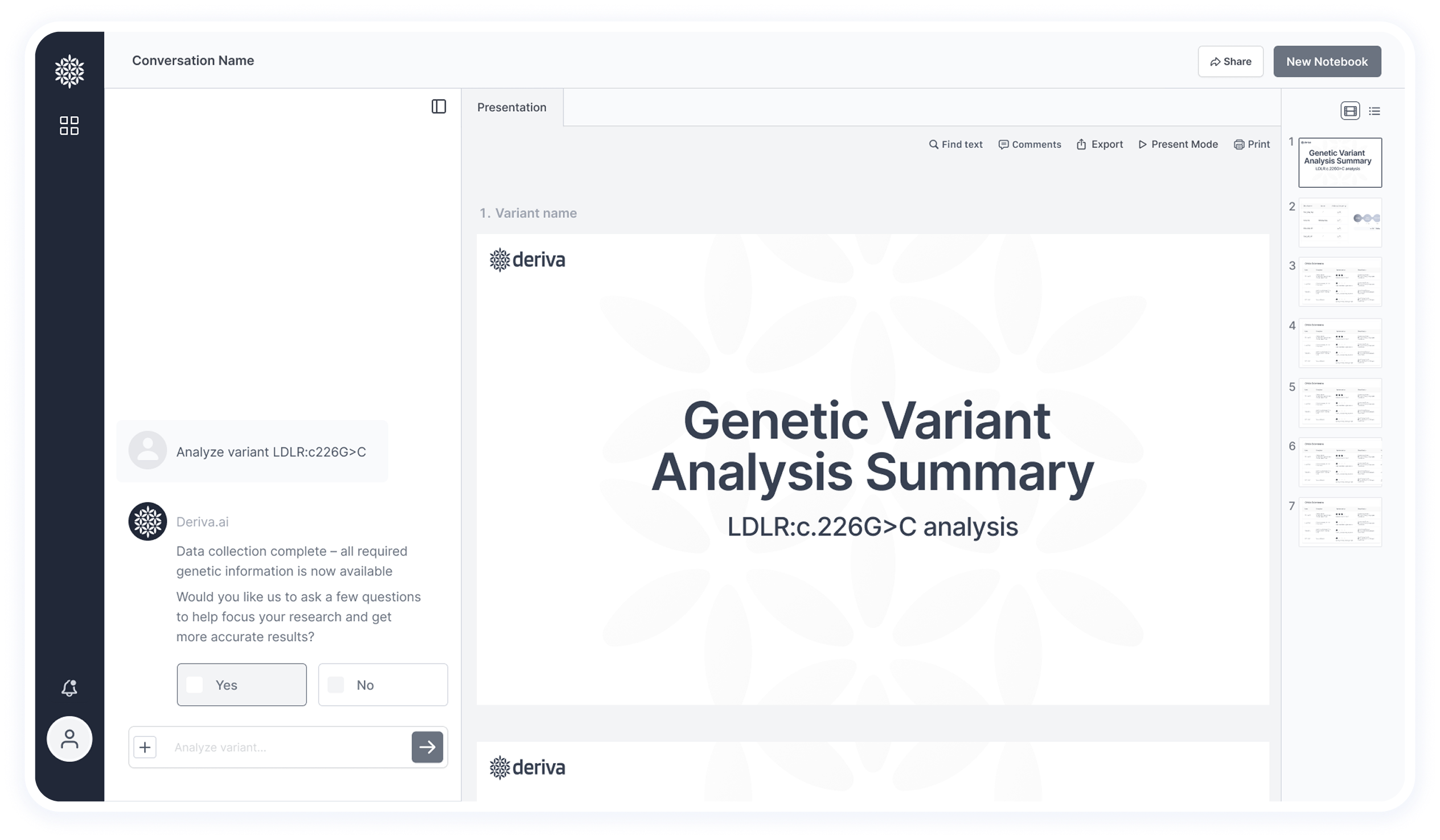
This screen shows the final results after data extraction and analysis — including insights and pathogenicity classification.
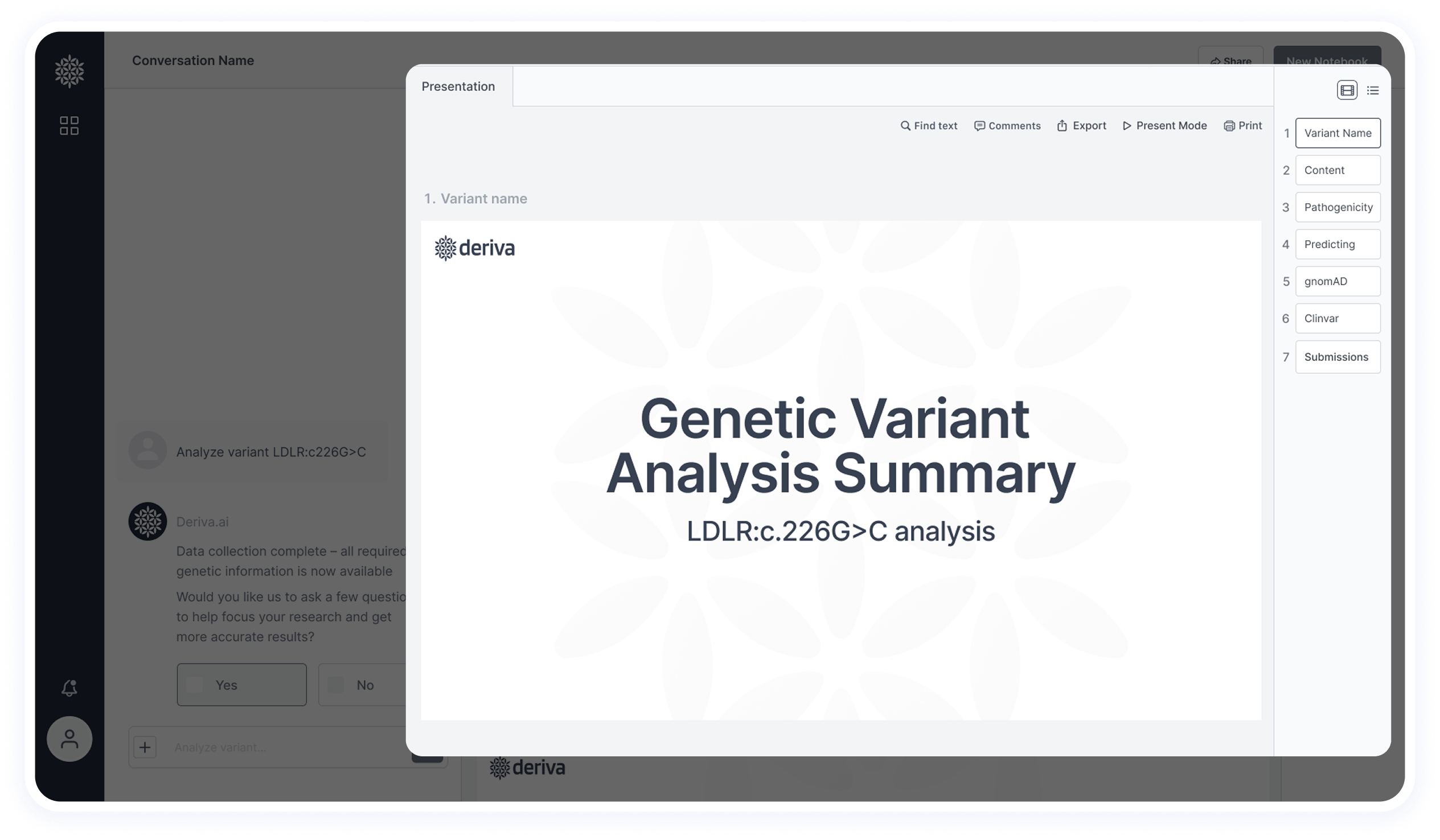
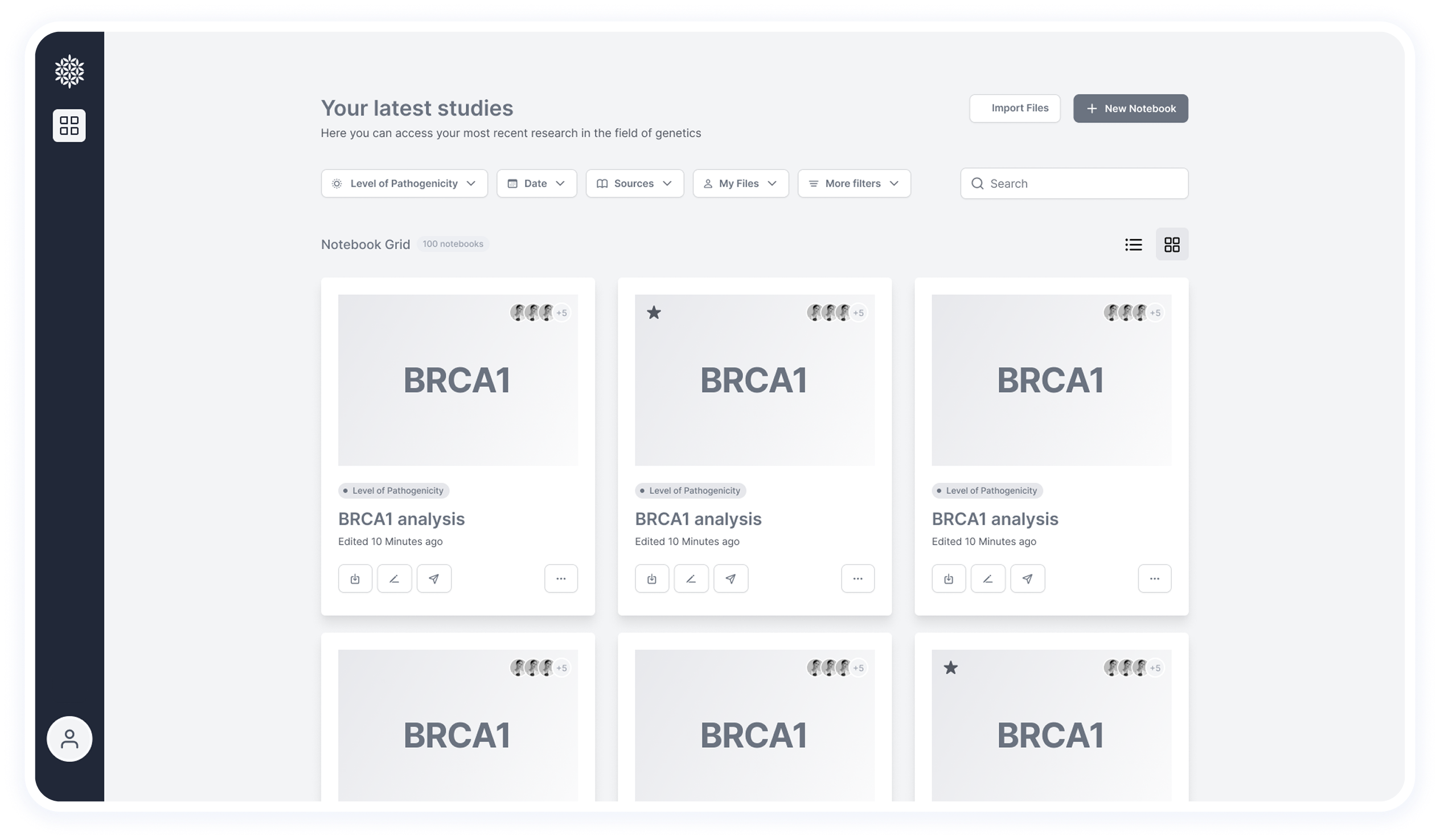
This section was simplified based on user feedback.
Originally, it included tabs for switching between multiple variants, but it created visual clutter and wasn’t aligned with researchers’ workflow — they usually focus on one variant at a time.
Now, the header includes essential actions like creating a new notebook and sharing with colleagues.
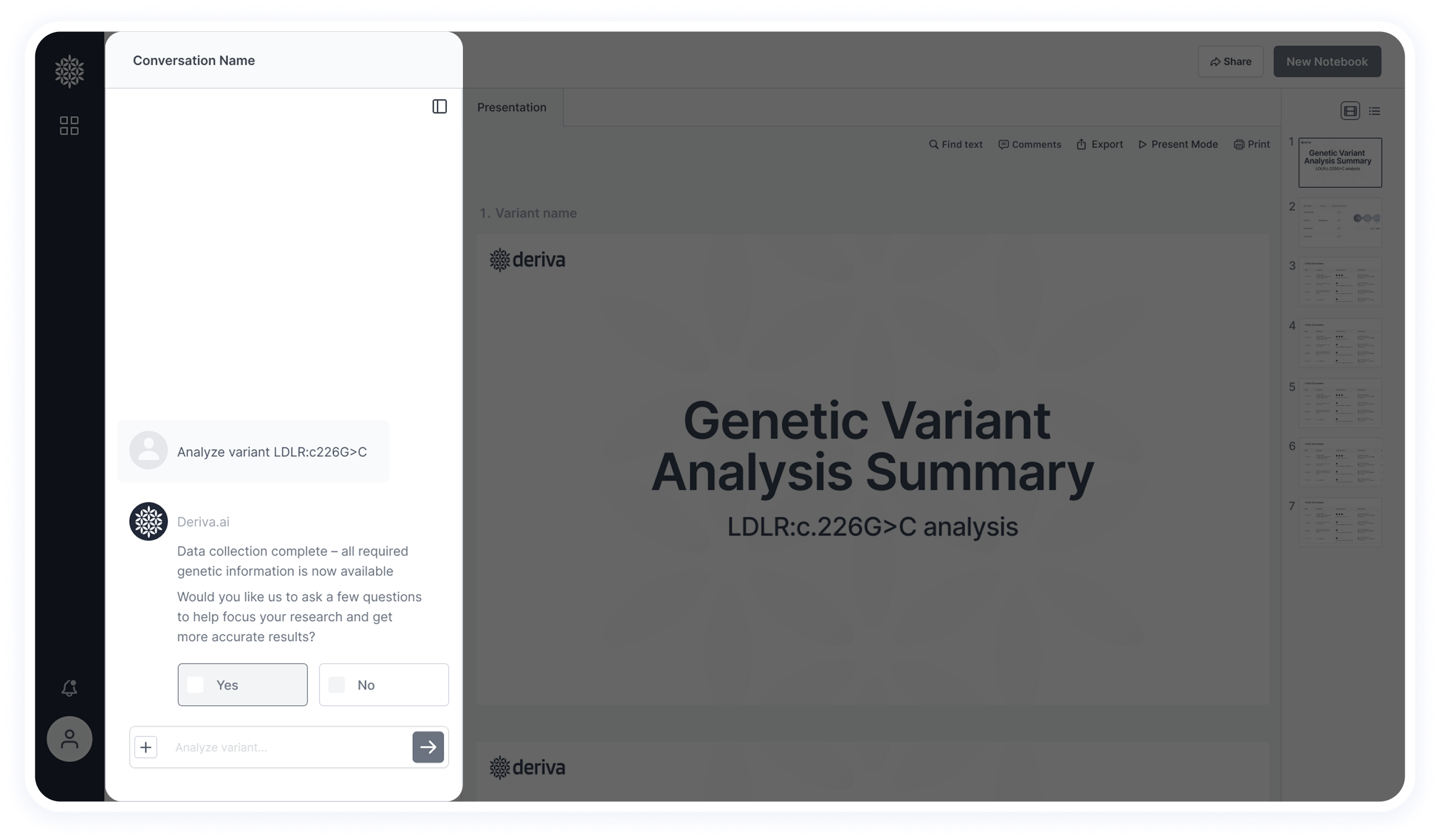
The chat combines open input with structured Yes/No questions to refine results and guide the research process more efficiently.
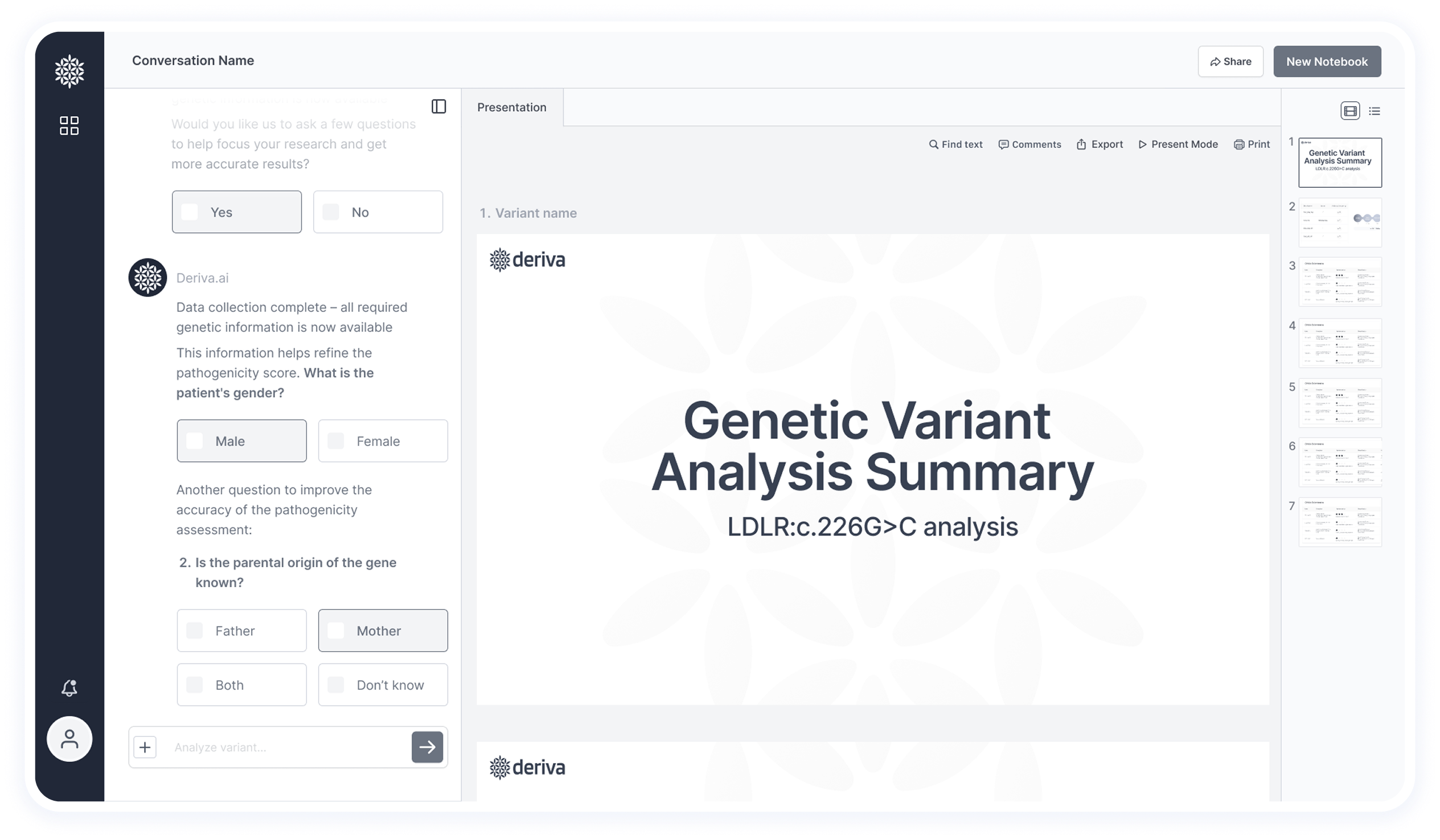
Some questions were identified as both automatable and essential for pathogenicity scoring — for example:
the patient’s sex, inheritance pattern (mother/father/both), and ethnic background.
These are provided directly through the chat as part of a streamlined, interactive experience.
- Users can highlight content and leave comments, with the option to tag colleagues in messages.
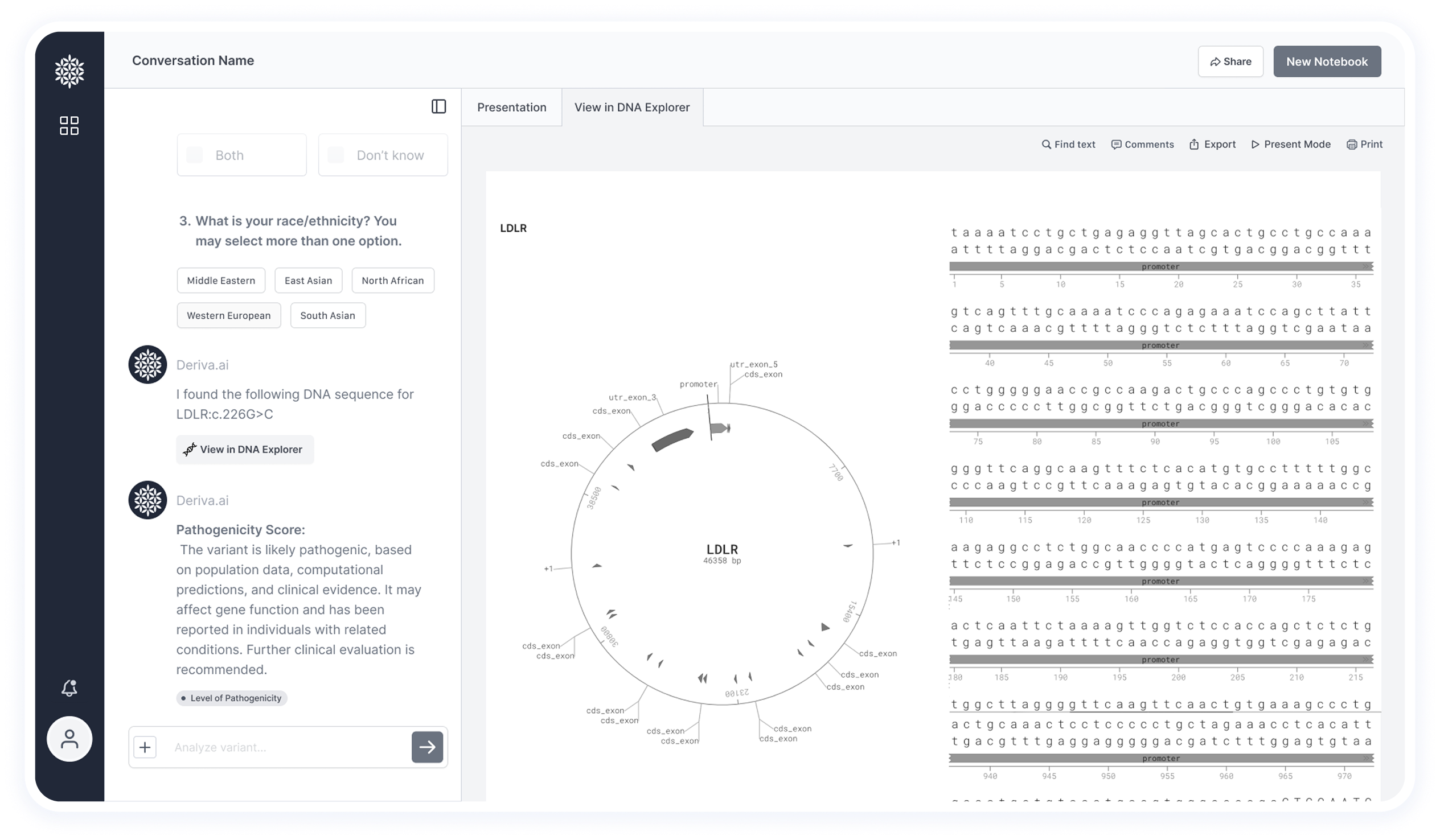
Certain research cases include important visual elements, such as protein structures.
These are displayed in a dedicated tab to support deeper scientific analysis.
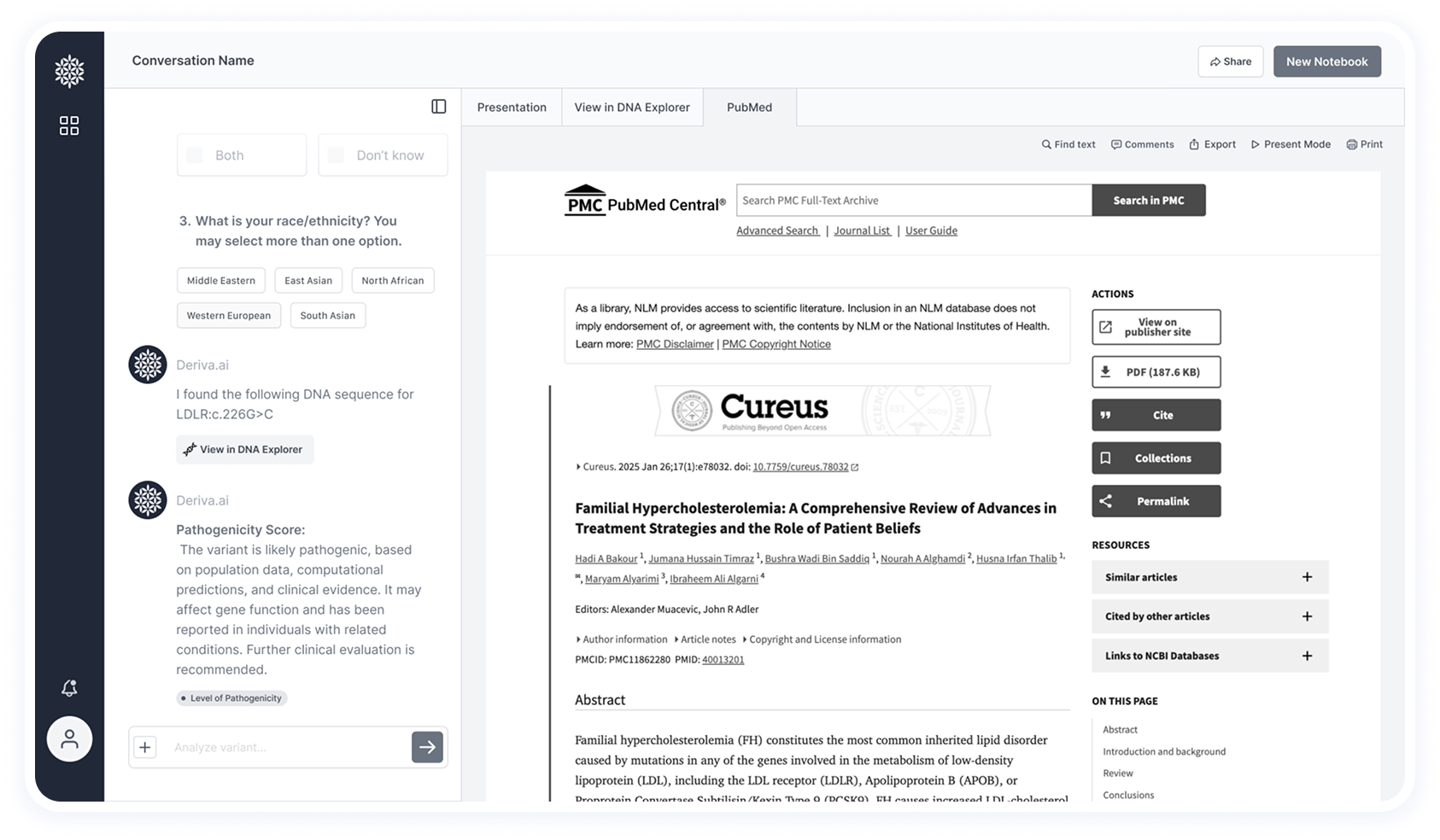
Deriva aims to keep the entire research workflow within the interface, without the need to switch between tools.
To support this, internal tabs are used to display all necessary content directly in the platform.
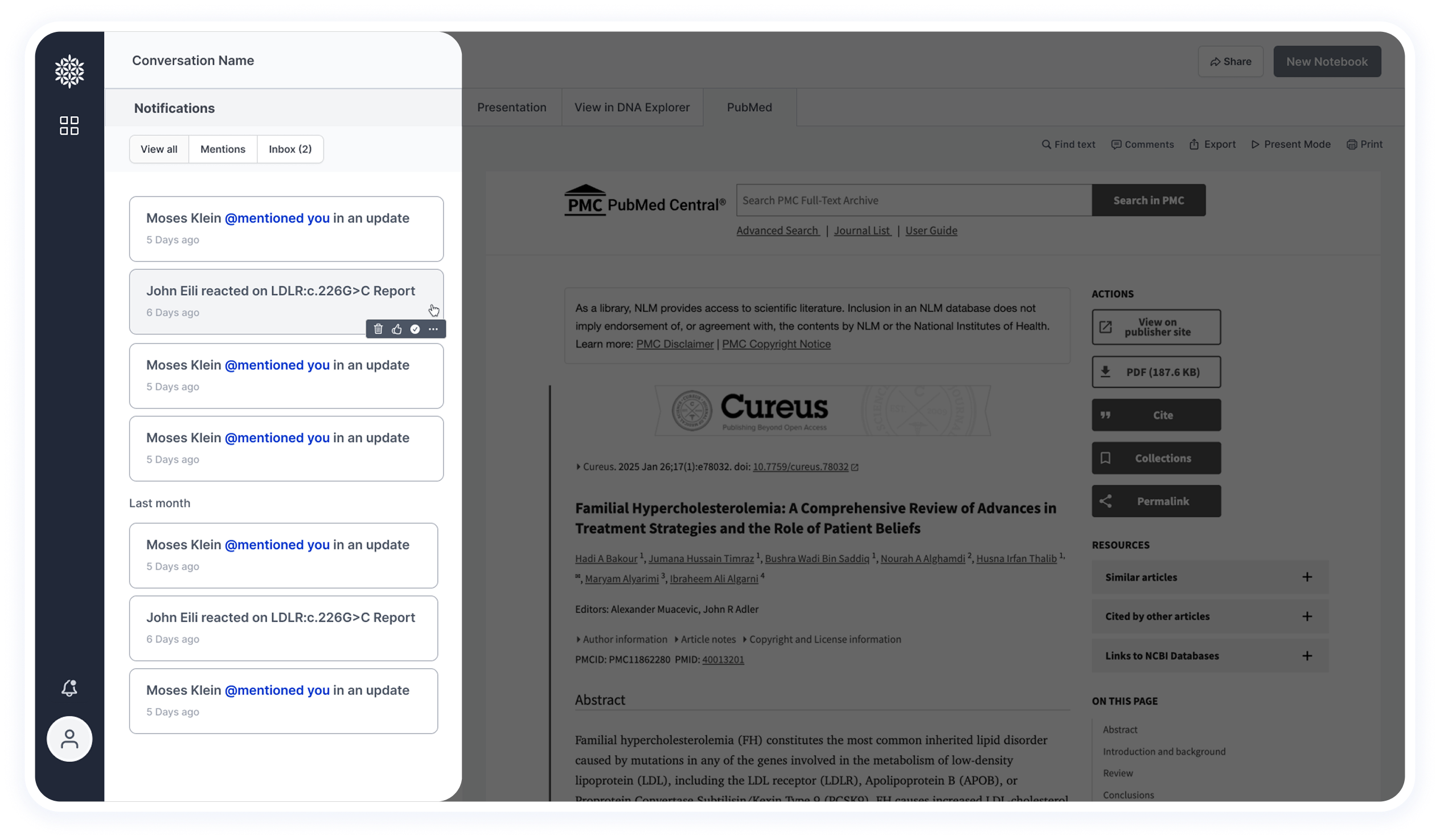
Based on insights from the user survey, notifications are highly valuable for researchers.
They are used to alert users when new information becomes available or when another researcher mentions them in a shared study.